The BReAst Carcinoma Subtyping (BRACS) is a new dataset of hematoxylin and eosin (H&E) histopathological images of breast carcinoma. BRACS has been built on the basis of an Agreement between IRCCS Fondazione Pascale, Institute for High Performance Computing and Networking (ICAR) of National Research Council (CNR), and IBM Research-Zurich for the “Development of methodologies and tools for the identification of atypical tumors in breast cancer pathology through the automatic analysis of histological images”.
This dataset offers a platform for researchers to compare strategies and algorithms for automated detection/classification of breast tumors in H&E stained tissue samples collected by mastectomy or biopsy. BRACS differs from most of the public breast cancer image datasets since it includes images representing atypical lesions. An early diagnosis of these atypical lesions could prevent the worsening into malignant cancer. In details, BRACS contains images characterized by the following kind of lesions: Pathological Benign (PB), Usual Ductal Hyperplasia (UDH), Flat Epithelial Atypia (FEA), Atypical Ductal Hyperplasia (ADH), Ductal Carcinoma in Situ (DCIS) and Invasive Carcinoma (IC). Also images representing Normal (N) tissue samples, i.e. glandular tissue samples without lesions, are included into BRACS.
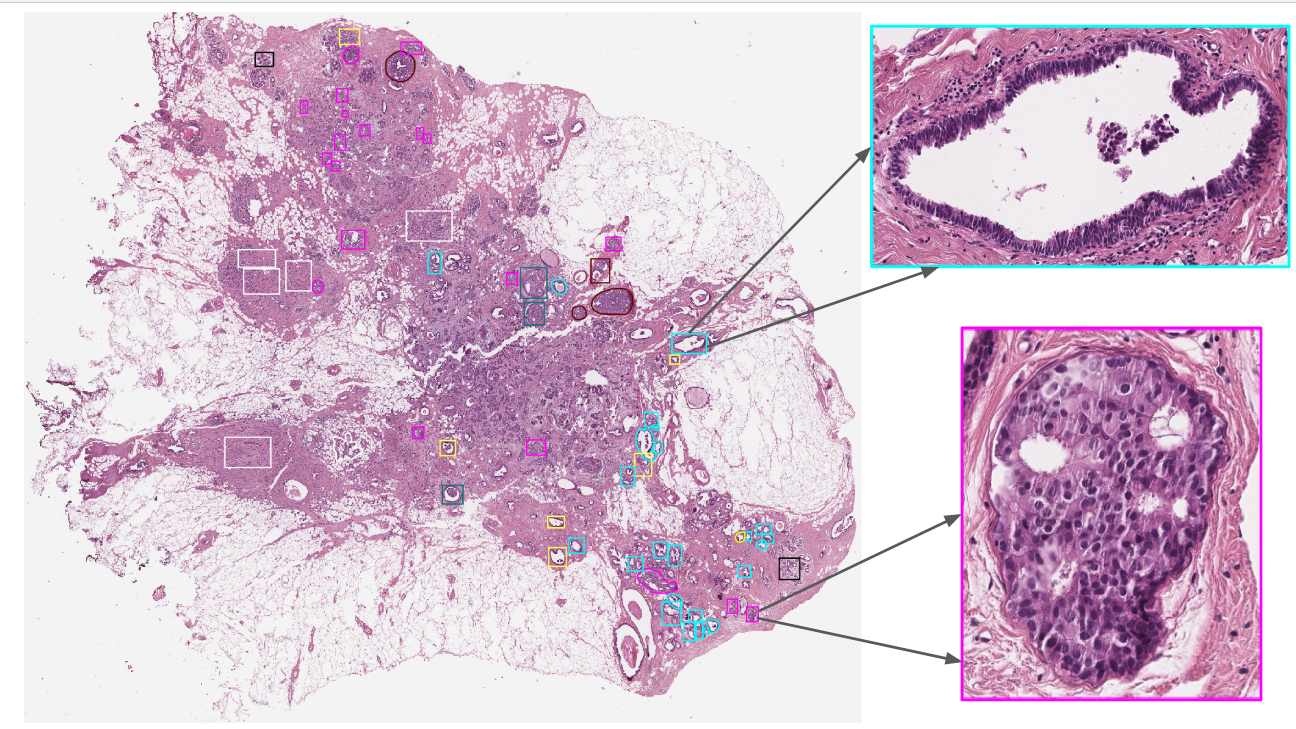
The Whole Slide
Images (WSI) of hematoxylin and eosin (H&E) stained breast tissues were generated by using an Aperio AT2 scanner at 0.25 µm/pixel for 40× resolution. Some Regions of Interest (RoIs) are associated with a subset of WSIs. See example figure below. Both WSIs and RoIs were annotated according to the seven classes mentioned above (N, PB, UDH, FEA, ADH, DCIS, IC), by three expert pathologists of the Complex Structure Pathological Anatomy and Cytopathology of National Cancer Institute – IRCCS Fondazione Pascale, Naples, Italy.
BRACS contains 547 WSIs collected by 189 patients and also includes 4537 RoIs acquired from 387 WSI collected by 151 patients. The WSIs (RoIs) do not have a fixed dimension and can easily exceed 100,000 by 100,000 (4,000 by 4,000) pixels.
If the results of algorithms running on BRACS datasets are to be used in scientific publications (journal publications, conference papers, technical reports, presentations at conferences and meetings) you should cite the following paper: “Brancati, N., Anniciello, A. M., Pati, P., Riccio, D., Scognamiglio, G., Jaume, G., De Pietro G., Di Bonito M., Foncubierta A., Botti G., Gabrani M., Feroce F. and Frucci, M. (2022). “BRACS: A dataset for breast carcinoma subtyping in H&E histology images”, Database, pp. 1-10.
 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. 
